In molecular biology research, DNA sequence analysis is the basis for further research and transformation of genes of interest. The current techniques for DNA sequencing are mainly the Sanger's Chain Termination Method, invented by Frederick Sanger. The advent of rapid DNA sequencing methods has greatly facilitated research and discovery in biology and medicine.
Sequencing principle
A DNA polymerase is used to extend the primer bound to the template of the sequence to be determined until a chain termination nucleotide is incorporated. Each sequence consists of a set of four separate reactions, each containing all four deoxynucleotide triphosphates (dNTPs) and spiked with a different amount of a different dideoxynucleoside triphosphate (ddNTP). Since the ddNTP lacks the 3-OH group required for extension, the extended oligonucleotide is selectively terminated at G, A, T or C. The termination point is determined by the corresponding dideoxy in the reaction. The relative concentrations of each of the dNTPs and ddNTPs can be adjusted to provide a set of chain termination products ranging from a few hundred to several kilobases in length. They have a common starting point, which is determined by the corresponding dideoxy in the reaction.
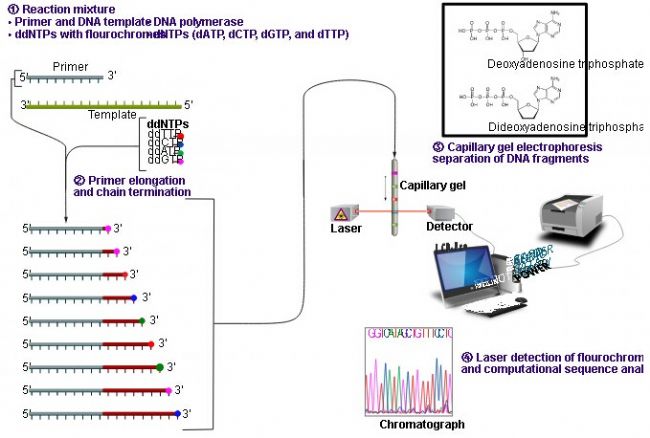
Sequencing method
The gene analyzer (ie, DNA sequencer) replaces the traditional polyacrylamide plate electrophoresis with capillary electrophoresis, and uses four-color fluorescent dye-labeled ddNTP (labeled terminator method), so the PCR product is generated by single primer PCR sequencing reaction. It is a single-stranded DNA mixture of 4 different fluorescent dyes at the 3' end of 1 base, so that the sequencing PCR products of the four fluorescent dyes can be electrophoresed in one capillary, thereby avoiding the influence of the difference in mobility between lanes. , greatly improving the accuracy of sequencing.
Due to the different molecular sizes, the mobility in capillary electrophoresis is also different. When passing through the capillary reading window segment, the CCD (charge-coupled device) camera detector in the laser detector window can detect the fluorescent molecules one by one. The fluorescence is spectrally split to distinguish the fluorescence of different colors representing different base information, and is simultaneously imaged on a CCD camera. The analysis software can automatically convert different fluorescence into DNA sequences for DNA sequencing purposes. The analysis results can be output in various forms such as gel electrophoresis pattern, fluorescence absorption peak map or base arrangement order.
Sequencing equipment
DNA sequencer (3730xl DNA Analyzer), performance characteristics, high degree of automation, providing continuous, no-monitoring operation, automatic filling, sample loading, electrophoresis separation, detection and data analysis, continuous operation for 24 hours without manual intervention. The sample is analyzed in a large amount, and a bundle of capillaries can perform full-automatic analysis on 96 samples at the same time, and can complete sequencing or fragment analysis of hundreds of samples in one day.
The advanced fluorescence detection system uses grating spectroscopic and CCD camera imaging technology to realize multi-color fluorescence simultaneous detection. Compared with the traditional filter and photomultiplier detection methods, the advantages of grating and CCD are that there is no need to replace any fluorescent chemistry. hardware equipment. The light emitted from the laser source is split into two by the optical element, and 96 capillary tubes are simultaneously irradiated from both sides by a laser beam, thereby effectively improving the sensitivity and uniformity of fluorescence detection.
Sequencing process
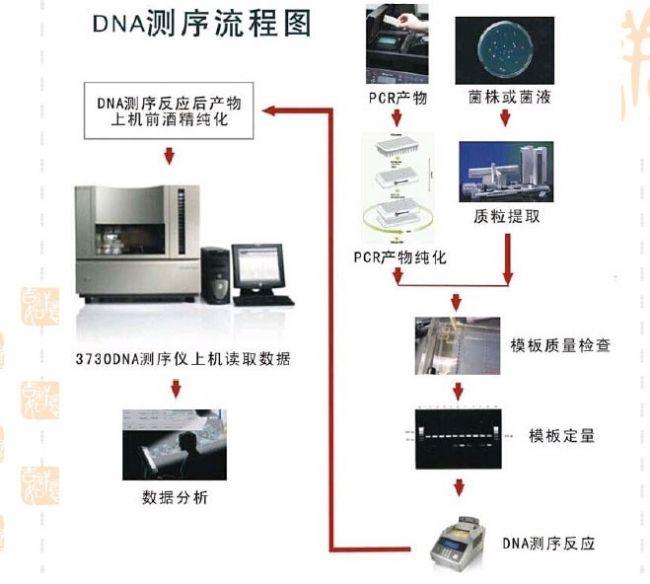
Guangzhou Zhongzhinan Supply Chain Co.,Ltd. , https://www.gzzhongzhinan.com